The genetic and epigenetic landscape of the Arabidopsis centromeres
Centromeres attach chromosomes to spindle microtubules during cell division and, despite this conserved role, show paradoxically rapid evolution and are typified by complex repeats. We used long-read sequencing to generate the Col-CEN Arabidopsis thaliana genome assembly that resolves all five centromeres. The centromeres consist of megabase-scale tandemly repeated satellite arrays, which support CENTROMERE SPECIFIC HISTONE H3 (CENH3) occupancy and are densely DNA methylated, with satellite variants private to each chromosome. CENH3 preferentially occupies satellites that show the least amount of divergence and occur in higher-order repeats. The centromeres are invaded by ATHILA retrotransposons, which disrupt genetic and epigenetic organization. Centromeric crossover recombination is suppressed, yet low levels of meiotic DNA double-strand breaks occur that are regulated by DNA methylation. We propose that Arabidopsis centromeres are evolving through cycles of satellite homogenization and retrotransposon-driven diversification.(Science)
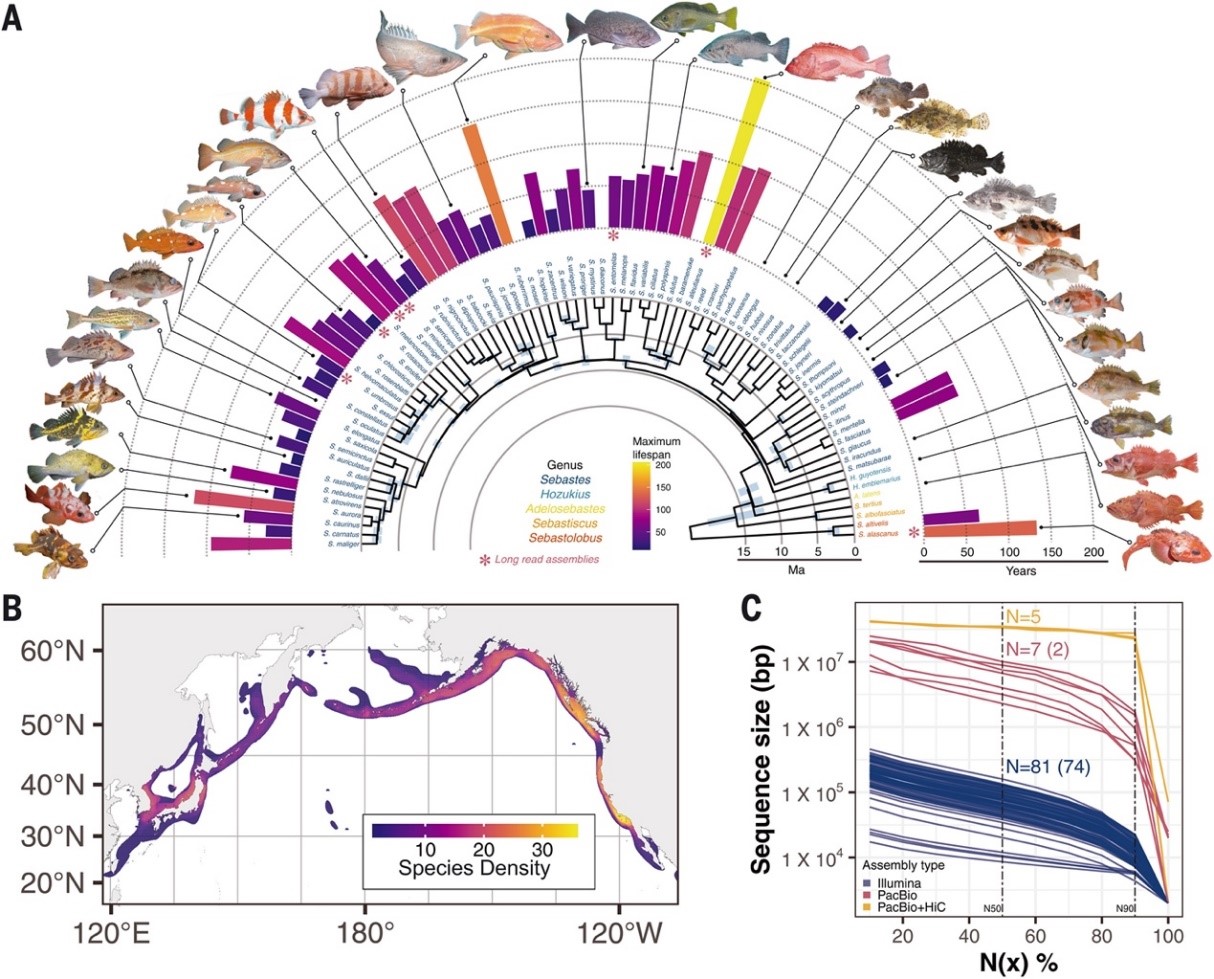
Origins and evolution of extreme life span in Pacific Ocean rockfishes
Pacific Ocean rockfishes (genus Sebastes) exhibit extreme variation in life span, with some species being among the most long-lived extant vertebrates. We de novo assembled the genomes of 88 rockfish species and from these identified repeated signatures of positive selection in DNA repair pathways in long-lived taxa and 137 longevity-associated genes with direct effects on life span through insulin signaling and with pleiotropic effects through size and environmental adaptations. A genome-wide screen of structural variation reveals copy number expansions in the immune modulatory butyrophilin gene family in long-lived species. The evolution of different rockfish life histories is coupled to genetic diversity and reshapes the mutational spectrum driving segregating CpG→TpG variants in long-lived species. These analyses highlight the genetic innovations that underlie life history trait adaptations and, in turn, how they shape genomic diversity.(Science )
A fishy tale of long and short life span
Fish have wide variations in life span even within closely related species. One such example are the rockfish species found along North Pacific coasts, which have life spans ranging from 11 to more than 200 years. Kolora et al. sequenced and performed a genomic analysis of 88 rockfish species, including long-read sequencing of the genomes of six species (see the Perspective by Lu et al.). From this analysis, the authors unmasked the genetic drivers of longevity evolution, including immunity and DNA repair–related pathways. Copy number expansion in the butyrophilin gene family was shown to be positively associated with life span, and population historical dynamics and life histories correlated differently between long- and short-lived species. These results support the idea that inflammation may modulate the aging process in these fish. —LMZ
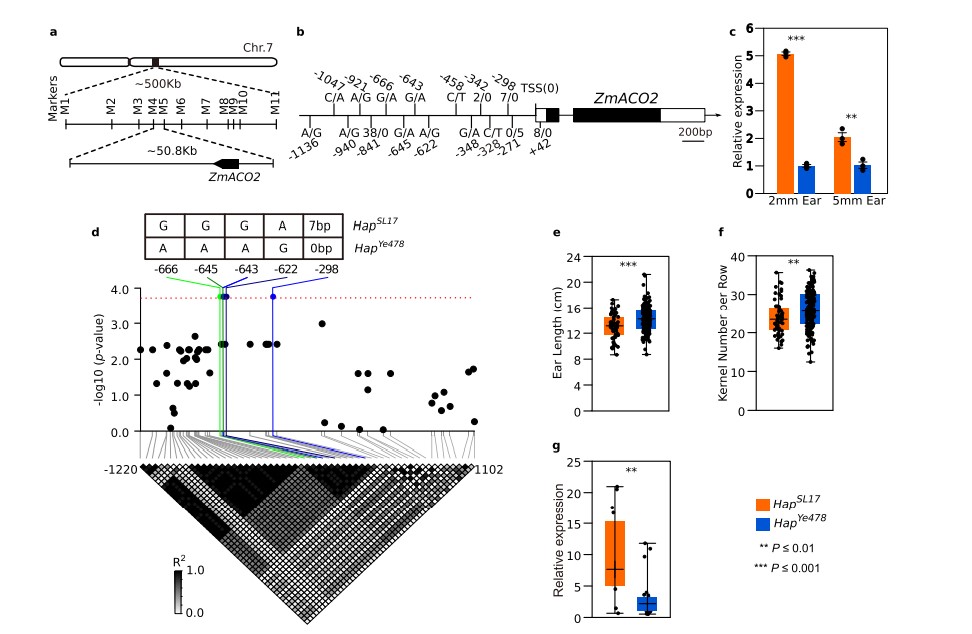
An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield
Maize ear size and kernel number differ among lines, however, little is known about the molecular basis of ear length and its impact on kernel number. Here, we characterize a quantitative trait locus, qEL7, to identify a maize gene controlling ear length, flower number and fertility. qEL7 encodes 1-aminocyclopropane-1-carboxylate oxidase2 (ACO2), a gene that functions in the final step of ethylene biosynthesis and is expressed in specific domains in developing inflorescences. Confirmation ofqEL7by gene editing ofZmACO2 leads to a reduction in ethylene production in developing ears, and promotes meristem and flower development, resulting in a ~13.4% increase in grain yield per ear in hybrids lines. Our findings suggest that ethylene serves as a key signal in inflorescence development, affecting spikelet number, floral fertility, ear length and kernel number, and also provide a tool to improve grain productivity by optimizing ethylene levels in maize or in other cereals.(Nature communications)

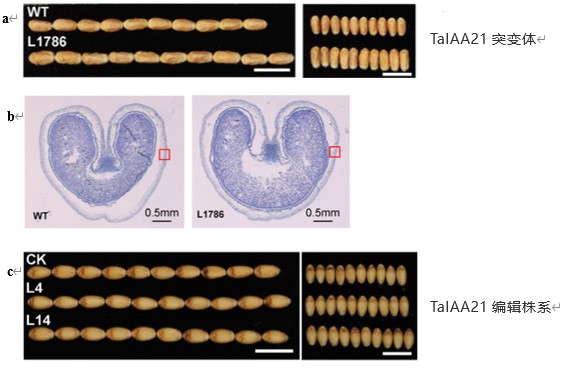
TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat
Auxin signaling is essential for the development of grain size and grain weight, two important components for crop yield. However, no auxin/indole acetic acid repressor (Aux/IAA) has been functionally characterized to be involved in the development of wheat (Triticum aestivum L.) grains to date. Here, we identified a wheat Aux/IAA gene, TaIAA21, and studied its regulatory pathway. We found that TaIAA21 mutation significantly increased grain length, grain width, and grain weight. Cross-sections of mutant grains revealed elongated outer pericarp cells compared to those of the wild type, where the expression of TaIAA21 was detected by in situ hybridization. Screening of auxin response factor (ARF) genes highly expressed in early developing grains revealed that TaARF25 interacts with TaIAA21. In contrast, mutation of the tetraploid wheat (Triticum turgidum) ARF25 gene significantly reduced grain size and weight. RNA sequencing analysis revealed upregulation of several ethylene response factor genes (ERFs) in taiaa21 mutants which carried auxin response cis-elements in their promoter. One of them, ERF3, was upregulated in the taiaa21 mutant and downregulated in the ttarf25 mutant. Transactivation assays showed that ARF25 promotes ERF3 transcription, while mutation of TtERF3 resulted in reduced grain size and weight. Analysis of natural variations identified three TaIAA21-A haplotypes with increased allele frequencies in cultivars relative to landraces, a signature of breeding selection. Our work demonstrates that TaIAA21 works as a negative regulator of grain size and weight development via the ARF25–ERFs module and is useful for yield improvement in wheat.(The Plant Journal)
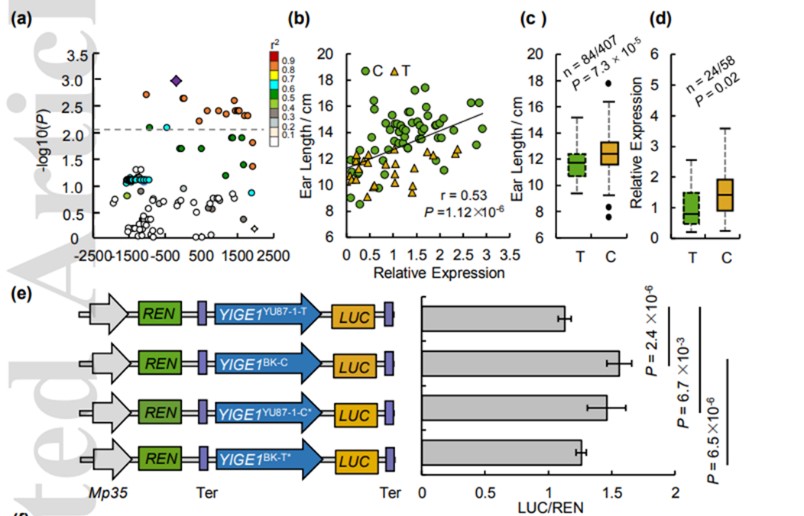
Genetic variation in YIGE1 contributes to ear length and grain yield in maize
• Ear length (EL), controlled by quantitative trait loci (QTL), is an important component of grain yield and as such a key target trait in maize breeding. However, very few EL QTL have been cloned and their molecular mechanisms are largely unknown. • Here, using genome wide association study (GWAS), we identified a QTL-YIGE1, which encodes an unknown protein that regulates EL by affecting pistillate floret number. Overexpression of YIGE1 enlarged female inflorescence meristem (IM) size, increased EL and kernel number per row (KNPR) thus enhanced grain yield. By contrast, CRISPR/Cas9 knock out or Mutator insertion mutant lines of YIGE1 displayed decreased IM size and EL. • The SNP located in the regulatory region of YIGE1 had strong effect on its promoter strength which positively affected EL by increasing gene expression. Further analysis shows that YIGE1 may be involved in sugar and auxin signal pathways to regulate maize ear development, thus, affecting IM activity and floret production in maize inflorescence morphogenesis. • These findings provide new insights of ear development and will ultimately facilitate maize molecular breeding.(New phytologist )
An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield
Maize ear size and kernel number differ among lines, however, little is known about the molecular basis of ear length and its impact on kernel number. Here, we characterize a quantitative trait locus, qEL7, to identify a maize gene controlling ear length, flower number and fertility. qEL7 encodes 1-aminocyclopropane-1- carboxylate oxidase2 (ACO2), a gene that functions in the final step of ethylene biosynthesis and is expressed in specific domains in developing inflorescences. Confirmation of qEL7 by gene editing of ZmACO2 leads to a reduction in ethylene production in developing ears, and promotes meristem and flower development, resulting in a ~13.4% increase in grain yield per ear in hybrids lines. Our findings suggest that ethylene serves as a key signal in inflorescence development, affecting spikelet number, floral fertility, ear length and kernel number, and also provide a tool to improve grain productivity by optimizing ethylene levels in maize or in other cereals.(Nature Communications)
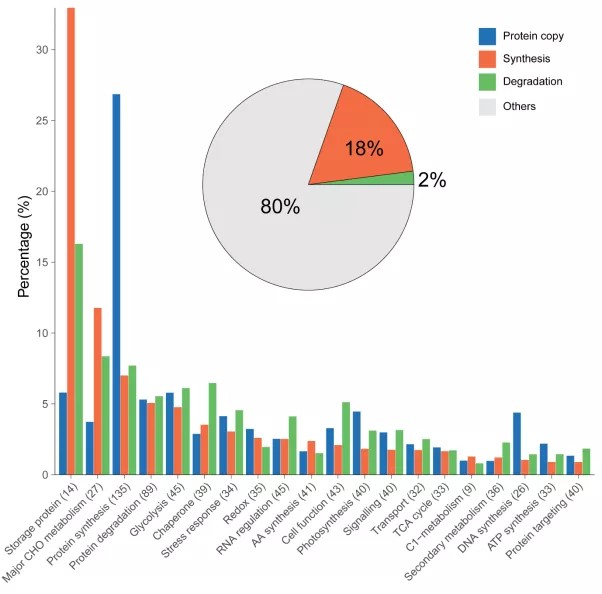
Protein turnover in the developing Triticum aestivum grain
Protein abundance in cereal grains is determined by the relative rates of protein synthesis and protein degradation during grain development but quantitation of these rates is lacking. Through combining in vivo stable isotope labelling and in-depth quantitative proteomics, we have measured the turnover of 1400 different types of proteins during wheat grain development. We demonstrate that there is a spatiotemporal pattern to protein turnover rates which explain part of the variation in protein abundances that is not attributable to differences in wheat gene expression. We show that c. 20% of total grain adenosine triphosphate (ATP) production is used for grain proteome biogenesis and maintenance, and nearly half of this budget is invested exclusively in storage protein synthesis. We calculate that 25% of newly synthesized storage proteins are turned over during grain development rather than stored. This approach to measure protein turnover rates at proteome scale reveals how different functional categories of grain proteins accumulate, calculates the costs of protein turnover during wheat grain development and identifies the most and the least stable proteins in the developing wheat grain.(New Phytologist)
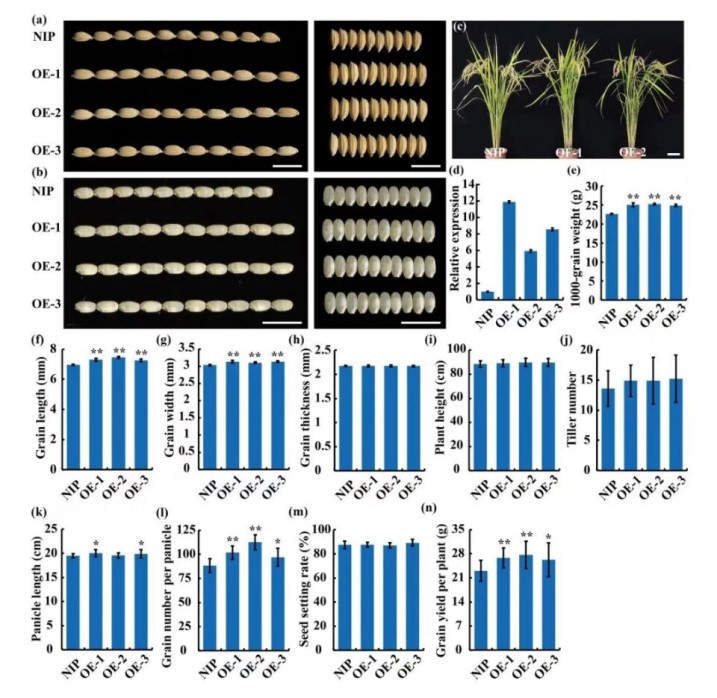
Plastidic pyruvate dehydrogenase complex E1 componentsubunit Alpha1 is involved in galactolipid biosynthesisrequired for amyloplast development in rice
Starch accounts for over 80% of the total dry weight in cereal endosperm and determines the kernel texture and nutritional quality. Amyloplasts, terminally differentiated plastids, are responsible for starch biosynthesis and storage. We screened a series of rice mutants with floury endosperm to clarify the mechanism underlying amyloplast development and starch synthesis.We identified the floury endosperm19(flo19) mutant which shows opaque of the interior endosperm. Abnormal compound starch grains (SGs) were present in the endosperm cells of the mutant. Molecular cloning revealed that theFLO19allele encodes a plastid-localized pyruvate dehydrogenase complex E1 component subunita1 (ptPDC-E1-a1) that is expressed in all rice tissues.In vivo enzyme assays demonstrated that theflo19mutant showed decreased activity of the plastidic pyruvate dehydrogenase complex. In addition, the amounts of monogalactosyl-diacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) were much lower in the developingflo19mutant endosperm, suggesting thatFLO19participates in fatty acid supply for galactolipid biosynthesis in amyloplasts.FLO19overexpression significantly increased seed size and weight,but did not affect other important agronomic traits, such as panicle length, tiller number andseed setting rate. An analysis of single nucleotide polymorphism data from a panel of rice accessions identified that thepFLO19Lhaplotype was positively associated with grain length,implying a potential application in rice breeding. In summary, our study demonstrates thatFLO19is involved in galactolipid biosynthesis which is essential for amyloplast development and starch biosynthesis in rice.(Plant Biotechnology Journal)
Protein turnover in the developing Triticum aestivum grain
Protein abundance in cereal grains is determined by the relative rates of protein synthesis and protein degradation during grain development but quantitation of these rates is lacking. Through combining in vivo stable isotope labelling and in-depth quantitative proteomics, we have measured the turnover of 1400 different types of proteins during wheat grain development. We demonstrate that there is a spatiotemporal pattern to protein turnover rates which explain part of the variation in protein abundances that is not attributable to differences in wheat gene expression. We show that c. 20% of total grain adenosine triphosphate (ATP) production is used for grain proteome biogenesis and maintenance, and nearly half of this budget is invested exclusively in storage protein synthesis. We calculate that 25% of newly synthesized storage proteins are turned over during grain development rather than stored. This approach to measure protein turnover rates at proteome scale reveals how different functional categories of grain proteins accumulate, calculates the costs of protein turnover during wheat grain development and identififies the most and the least stable proteins in the developing wheat grain.(New phytologist)
The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use Efficiency
The role of abscisic acid (ABA) receptors, PYR1/PYL/RCAR (PYLs), is well established in ABA signalling and plant drought response, but limited research has explored the regulation of wheat PYLs in this process, especially the effects of their allelic variations on drought tolerance or grain yield. Here, we found that the overexpression of a TaABFs-regulated PYL gene, TaPYL1-1B, exhibited higher ABA sensitivity, photosynthetic capacity and water-use effificiency (WUE), all contributed to higher drought tolerance than that of wild-type plants. This heightened watersaving mechanism further increased grain yield and protected productivity during water defificit. Candidate gene association analysis revealed that a favourable allele TaPYL1-1BIn-442, carrying an MYB recognition site insertion in the promoter, is targeted by TaMYB70 and confers enhanced expression of TaPYL1-1B in drought-tolerant genotypes. More importantly, an increase in frequency of the TaPYL1-1BIn-442 allele over decades among modern Chinese cultivars and its association with high thousand-kernel weight together demonstrated that it was artifificially selected during wheat improvement efforts. Taken together, our fifindings illuminate the role of TaPYL1-1B plays in coordinating drought tolerance and grain yield. In particular, the allelic variant TaPYL1-1BIn-442 substantially contributes to enhanced drought tolerance while maintaining high yield, and thus represents a valuable genetic target for engineering droughttolerant wheat germplasm.(Plant Biotechnology Journal)
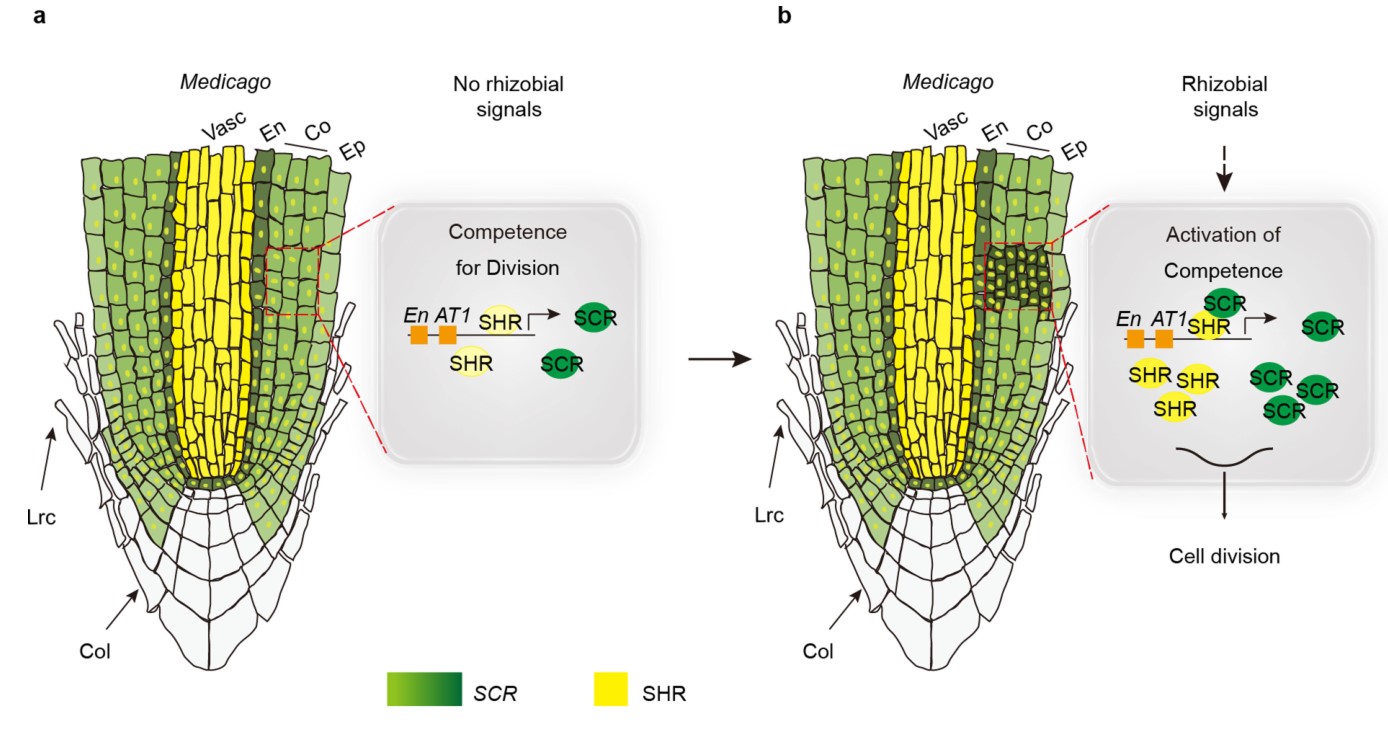
An SHR–SCR module specifies legume cortical cell fate to enable nodulation
Legumes, unlike other plants, have the ability to establish symbiosis with nitrogen-fixing rhizobia. It has been theorized that a unique property of legume root cortical cells enabled the initial establishment of rhizobial symbiosis 1–3 . Here we show that a SHORTROOT SCARECROW (SHR–SCR) stem cell program in cortical cells of the legume Medicago truncatula specifies their distinct fate. Regulatory elements drive the cortical expression of SCR, and stele-expressed SHR protein accumulates in cortical cells of M. truncatula but not Arabidopsis thaliana. The cortical SHR–SCR network is conserved across legume species, responds to rhizobial signals, and initiates legume-specific cortical cell division for de novo nodule organogenesis and accommodation of rhizobia. Ectopic activation of SHR and SCR in legumes is sufficient to induce root cortical cell division. Our work suggests that acquisition of the cortical SHR–SCR module enabled cell division coupled to rhizobial infection in legumes. We propose that this event was central to the evolution of rhizobial endosymbiosis.(Nature)
The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency
The role of abscisic acid (ABA) receptors, PYR1/PYL/RCAR (PYLs), is well established in ABA signaling and plant drought response, but limited research has explored the regulation of wheat PYLs in this process, especially the effects of their allelic variations on drought tolerance or grain yield. Here, we found that overexpression of a TaABFs-regulated PYL gene, TaPYL1-1B, exhibited higher ABA sensitivity, photosynthetic capacity, and water-use efficiency (WUE), all contributed to higher drought tolerance than that of wild-type plants. This heightened water-saving mechanism further increased grain yield and protected productivity during water deficit. Candidate gene association analysis revealed that a favorable allele TaPYL1-1B In-442, carrying an MYB recognition site insertion in the promoter, is targeted by TaMYB70 and confers enhanced expression of TaPYL1-1B in drought-tolerant genotypes. More importantly, an increase in frequency of the TaPYL1-1B In-442 allele over decades among modern Chinese cultivars and its association with high thousand-kernel weight together demonstrated that it was artificially selected during wheat improvement efforts. Taken together, our findings illuminate the role of TaPYL1-1B plays in coordinating drought tolerance and grain yield. In particular, the allelic variant TaPYL1-1B In-442 substantially contributes to enhanced drought tolerance while maintaining high yield, and thus represents a valuable genetic target for engineering drought-tolerant wheat germplasm.(Plant Biotechnology Journal)
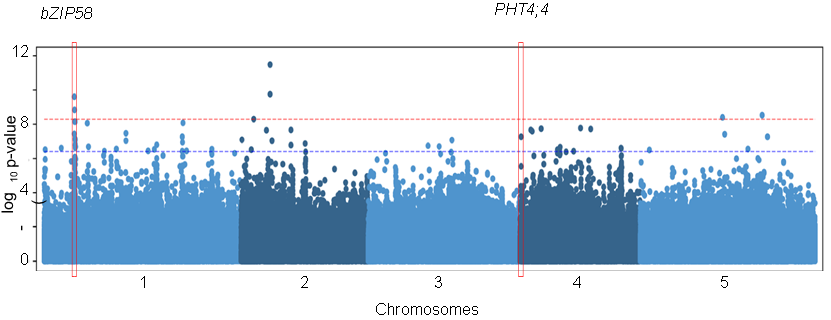
Interdependent iron and phosphorus availability controls photosynthesis through retrograde signaling
Iron deficiency hampers photosynthesis and is associated with chlorosis. We recently showed that iron deficiency-induced chlorosis depends on phosphorus availability. How plants integrate these cues to control chlorophyll accumulation is unknown. Here, we show that iron limitation downregulates photosynthesis genes in a phosphorus-dependent manner. Using transcriptomics and genome-wide association analysis, we identify two genes, PHT4;4 encoding a chloroplastic ascorbate transporter and bZIP58, encoding a nuclear transcription factor, which prevent the downregulation of photosynthesis genes leading to the stay-green phenotype under iron-phosphorus deficiency. Joint limitation of these nutrients induces ascorbate accumulation by activating expression of an ascorbate biosynthesis gene, VTC4, which requires bZIP58. Furthermore, we demonstrate that chloroplastic ascorbate transport prevents the downregulation of photosynthesis genes under iron-phosphorus combined deficiency through modulation of ROS homeostasis. Our study uncovers a ROS-mediated chloroplastic retrograde signaling pathway to adapt photosynthesis to nutrient availability.(Nature Communications)
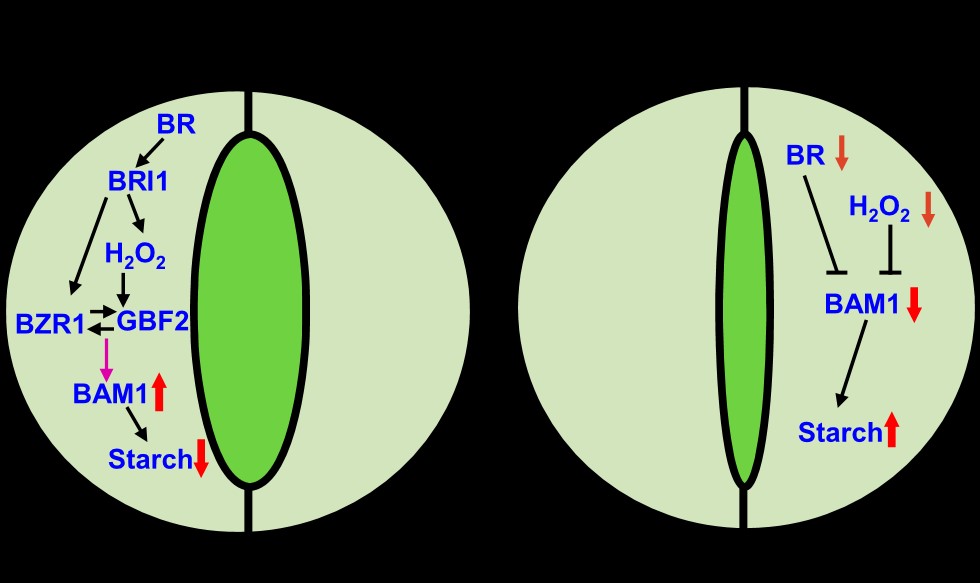
Brassinosteroid and Hydrogen Peroxide Interdependently Induce Stomatal Opening by Promoting Guard Cell Starch Degradation
Starch is the major storage carbohydrate in plants and functions in buffering carbon and energy availability for plant fitness with challenging environmental conditions. The timing and extent of starch degradation appear to be determined by diverse hormonal and environmental signals; however, our understanding of the regulation of starch metabolism is fragmentary. Here, we demonstrate that the phytohormone brassinosteroid (BR) and redox signal hydrogen peroxide (H2O2) induce the breakdown of starch in guard cells, which promotes stomatal opening. The BR-insensitive mutant bri1-116 accumulated high levels of starch in guard cells, impairing stomatal opening in response to light. The gain-of-function mutant bzr1-1D suppressed the starch excess phenotype of bri1-116, thereby promoting stomatal opening. BRASSINAZOLE-RESISTANT1 (BZR1) interacts with the basic leucine zipper transcription factor G-BOX BINDING FACTOR2 (GBF2) to promote the expression of β-AMYLASE1 (BAM1), which is responsible for starch degradation in guard cells. H2O2 induces BZR1 oxidation, enhancing the interaction between BZR1 and GBF2 to increase BAM1 transcription. Mutations in BAM1 lead to starch accumulation and reduce the effects of BR and H2O2 on stomatal opening. Overall, this study uncovers the critical roles of BR and H2O2 in regulating guard cell starch metabolism and stomatal opening(The Plant Cell)
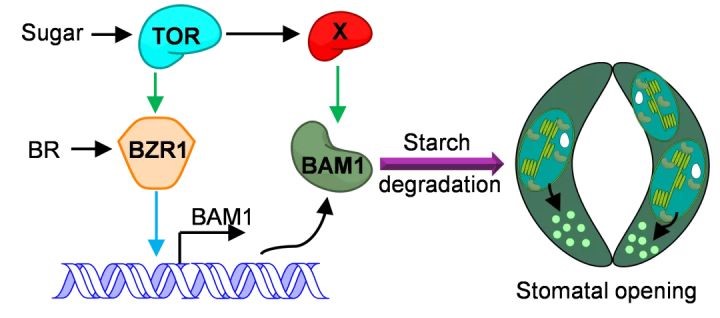
TOR promotes guard cell starch degradation by regulating the activity of β-AMYLASE1 in Arabidopsis
Starch is the main energy storage carbohydrate in plants and serves as an essential carbon storage molecule for plant metabolism and growth under changing environmental conditions. The TARGET of RAPAMYCIN (TOR) kinase is an evolutionarily conserved master regulator that integrates energy, nutrient, hormone, and stress signaling to regulate growth in all eukaryotes. Here, we demonstrate that TOR promotes guard cell starch degradation and induces stomatal opening in Arabidopsis thaliana. Starvation, caused by plants growing under short photoperiod or low light photon irradiance, as well as inactivation of TOR, impaired guard cell starch degradation and stomatal opening. Sugar and TOR induce the accumulation of β-AMYLASE1 (BAM1), which is responsible for starch degradation in guard cells. The plant steroid hormone brassinosteroid (BR) and transcription factor BZR1 play crucial roles in sugar-promoted expression of BAM1. Furthermore, sugar supply induced BAM1 accumulation, but TOR inactivation led to BAM1 degradation, and the effects of TOR inactivation on BAM1 degradation were abolished by the inhibition of autophagy and proteasome pathways or by phospho-mimicking mutation of BAM1 at serine-31. Such regulation of BAM1 activity by sugar–TOR signaling allows carbon availability to regulate guard cell starch metabolism and stomatal movement, ensuring optimal photosynthesis efficiency of plants.(The Plant Cell)