HebQTLs reveal intra-subgenome regulation inducing unbalanced expression and function among bread wheat homoeologs
Abstract
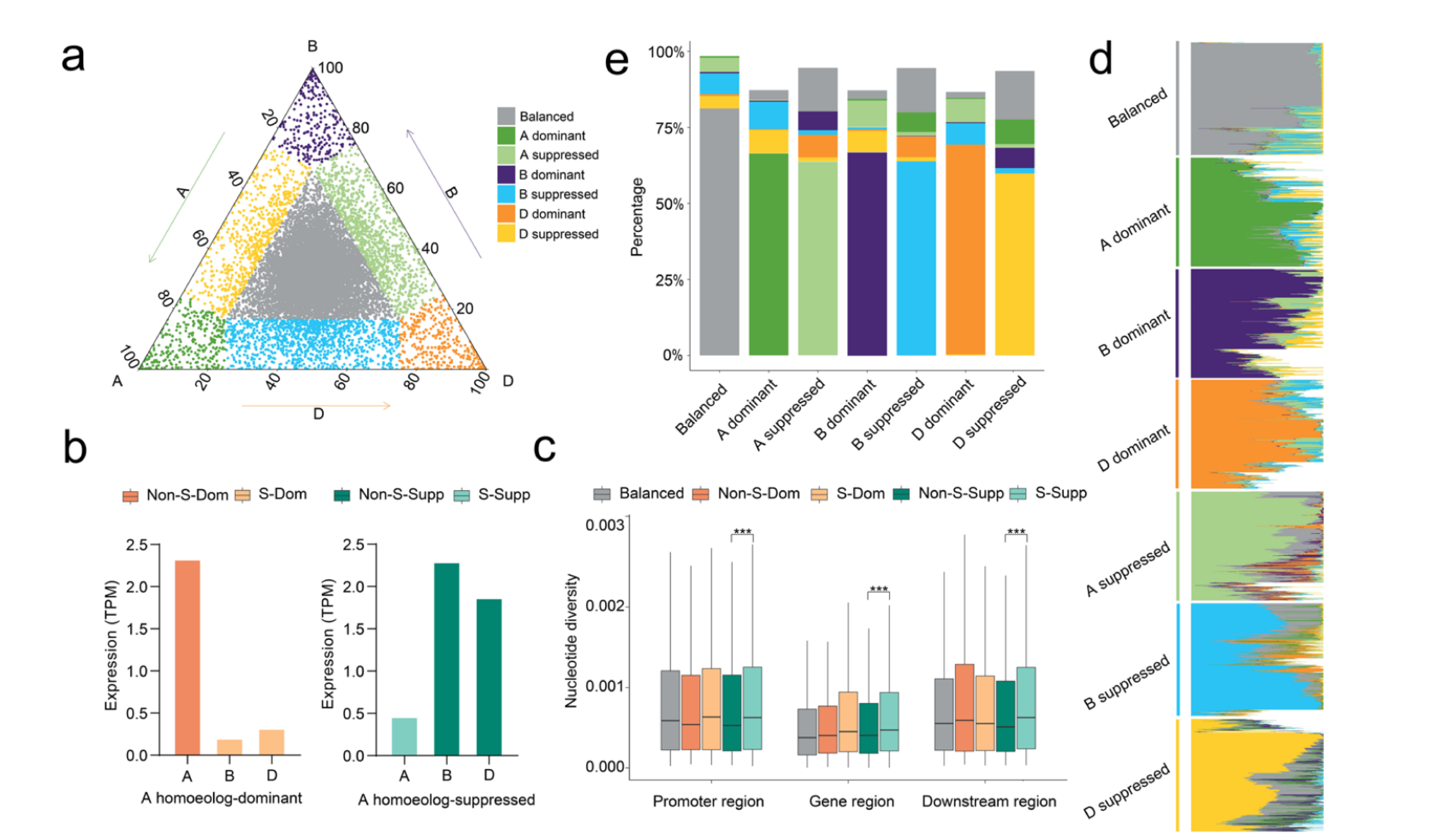
Here we uncover substantial variation in homoeolog expression bias among the root transcriptomes of a natural population of 406 bread wheat (Triticum aestivum ssp. aestivum) accessions collected worldwide. We develop a new model allowing us to identify 14,727 quantitative trait loci regulating the variation in homoeolog expression bias (hebQTLs), indicating that homoeolog expression bias is genetically regulated and can be predicted using genotyping data. The hebQTLs mostly regulate the expression of homoeologs in the same subgenome and downregulate expression to produce homoeolog expression bias, suggesting that intra-subgenomic rather than inter-subgenomic interactions induce homoeolog expression bias. Furthermore, we determine that hebQTL-regulated homoeologs exhibit higher genetic diversity and weaker biological functions than their counterparts. Notably, the downregulation of 38.4% of hebQTL-regulated homoeologs is compensated for by the upregulation of other homoeologs within the triad.
本站总访问量次 本站总访客数人 本文总阅读量次